HiD 3D Reconstruction
HistoDigital (HiD) 3D Reconstruction is currently offered as a service to enable the creation of 3D digital reconstructions of anatomical tissue structures from histological slide images.
In the process of 3D spatial reconstruction of your data with HiD, we attach particular importance to the anatomically accurate reconstruction of the original tissues through iterative application of algorithms specfically designed and crafted for this purpose.
Applications
- Digital Pathology
- Embryological and oncological research
- Education in anatomy and physiology
Solutions
- Assisted sorting and alignment
- Correction of staining variances
- Rigid and nonrigid slice alignment
- 3D volumetric reconstruction
Benefits
- Assess tissue from all viewing angles
- True 3D tissue analysis
- Exemption of anatomical structures
- Visualization of the topographical situation
- Breathtaking image quality
Understand the position and course of anatomical structures in tissue and analyze topography in full 3D context.
Technical Background of HiD 3D Reconstruction
In HiD 3D Reconstruction of anatomical slice data, we place particular emphasis on the anatomically accurate reconstruction of the original tissues through the iterative application of special algorithms to enable meaningful analysis and interpretation of the structures in 3D.
This is necessary because a wide variety of distortions can occur, for example during slicing, the placement on the slides, or the digitization.
We correct these distortions by using a range of innovative technologies, which we successfully apply to 3D reconstructions thanks to years of research and development together with HiD.
Slice Extraction
The selection of slides and the extraction of slices is the first step in the reconstruction workflow.
- Automatic detection of the slices on the slide
- Interactive options for sorting and correction
Intensity Correction
In this step, we correct intensity inhomogeneities between neighboring slices that may happen due to lighting effects in digitization or due to staining inconsistencies.
- Correction of intensity inhomogeneities
- Control of intensity variation curves along the slice stack
Reconstruction
The final stage reconstructs the full slice stack using rigid and nonrigid registration techniques, which account for slice placement and deformation.
- Rigid registration for slice placement correction
- Nonrigid registration to compensate slicing deformations
- Reconstruction into consistent 3D volume (MPRs, volume rendering)
3D Reconstruction Example
The following example shows some results from a 3D reconstruction of a fetal kidney dataset, which consists of roughly 850 slices.
The following interim results show the effect of image registration on the quality of cross-sectional context, which is best demonstrated by MPR (multiplanar reformatting) visualizations perpendicular to the slice stack (the right two images each).
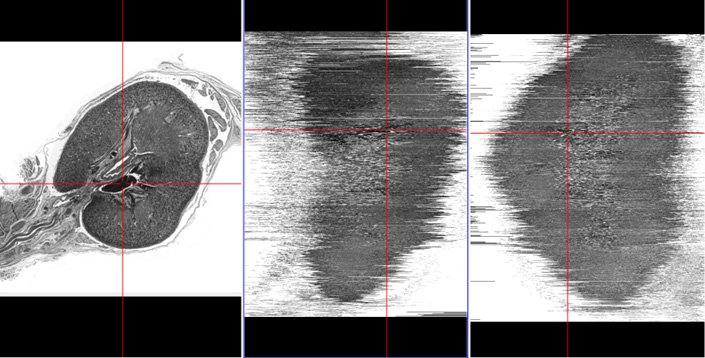
Slice reconstruction by simply placing the extracted slices onto each other (no registration).

Result after rigid registration of the slices in their respective neighboring context. Slice distortions still remain.
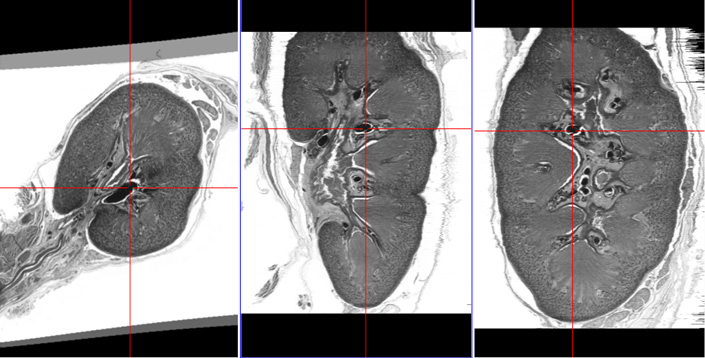
Final reconstruction after nonrigid registration. Details are now visible in full 3D context.
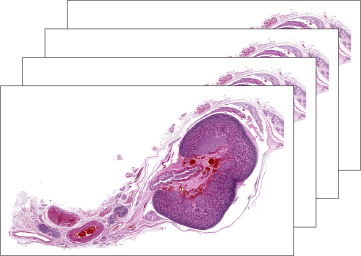
Original histological slices from a fetal kidney dataset.

Coronal (left) and sagittal (right) MPR (multiplanar reformatting) visualizations perpendicular to the slice stack after 3D reconstruction.
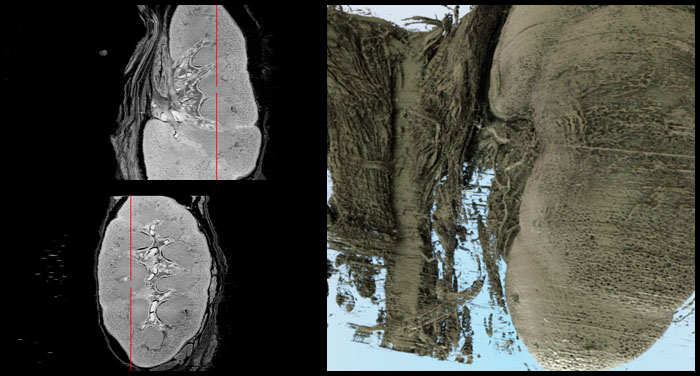

Visualizations of the kidney slices as a result of the HiD 3D Reconstruction.
Tissue Morphology in 3D
In order to understand the morphology in the tissue and to analyze the topographical situation in a specific area in detail, it is necessary to segment anatomical structures.
The results of the HiD 3D Reconstruction can be seamlessly imported and processed in the Chimaera Annotation Client for further processing.
HiD 3D Reconstruction thus offers a wide range of possibilities for 3D tissue analysis.


Reconstruction of a paraffin embedded pig lacrimal grand (grey) along with a segmentation of seven excretory lacrimal ducts (colored).
References
- Ali, Mohammad; Zetzsche, Maximilian; Scholz, Michael; Hahn, Dieter; Gaffling, Simone; Heichel, Jens; Hammer, Christian; Bräuer, Lars; Paulsen, Friedrich. "New insights into the lacrimal pump", The Ocular Surface, 18, 2020, 10.1016/j.jtos.2020.07.013
- Gaffling, Simone; Daum, Volker; Steidl, Stefan; Maier, Andreas; Köstler, Harald; Hornegger, Joachim. "A Gauss-Seidel Iteration Scheme for Reference-Free 3-D Histological Image Reconstruction", IEEE Transactions on Medical Imaging, vol. 34, no. 2, pp. 514-530, 2015
- Henker, Robert; Scholz, Michael; Gaffling, Simone; Asano, Nagayoshi; Hampel, Ulrike; Garreis, Fabian; Hornegger, Joachim; Paulsen, Friedrich. "Morphological Features of the Porcine Lacrimal Gland and Its Compatibility for Human Lacrimal Gland Xenografting", PLoS one, vol. 8, no. 9, pp. e74046, 2013
- Graffling, Simone; Daum, Volker; Hornegger, Joachim. "Landmark-constrained 3-D Histological Imaging: A Morphology-preserving Approach", VMV 2011: Vision, Modeling & Visualization (16th International Workshop on Vision, Modeling and Visualization 2011), Berlin, 04-06.10.2011, pp. 309-316, 2011, ISBN 978-3-905673-85-2
- Karen Lahme, Wiebke Sachs, Sarah Froembling, Desiree Loreth, Vincent Böttcher-Dierks, Katrin Neumann, Frederik-Michael Hann, Nick Arkan, Michael Brehler, Julia Reichelt, Antonia Sgries, Kristin Surmann, Simone Gaffling, Marie R. Adler, Pablo J. Sáez, Uta Wedekind, Alina Lampert, Elena Tasika, Paul Saftig, Christian Conze, …, Catherine Meyer-Schwesinger “Autoantibody-triggered podocyte membrane budding drives autoimmune kidney disease”, https://doi.org/10.1016/j.cell.2025.11.010

